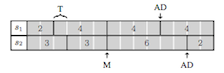
Developer Interface¶
The APIs for most metrics can be provided either two segmentations to compare or a dataset to perform pairwise comparisons upon. There are a variety of parameters that can be specified other than that which is compared, but all have defaults specified.
Boundary-Edit-Distance-based Metrics¶
These segmentation comparison metrics were introduced in [Fournier2013].
-
segeval.boundary_statistics(*args, **kwargs)¶ - Computes a large number of BED-based and other segmentation statistics, returning a
dict()that includes: count_edits, a count of BED edits;additions, a list of BED addition edits;substitutions, a list of BED substitution edits;transpositions, a list of BED transposition edits;full_misses, a list of fully-missed boundaries (regardless of edits);boundaries_all, a count of boundaries compared;matches, a list of matching boundaries;pbs, a count of potential boundary types.
- Computes a large number of BED-based and other segmentation statistics, returning a
-
class
segeval.BoundaryFormat¶ - An
enumwith options that include: sets, a boundary set string; seeboundary_string_from_masses()mass, a tuple of segment masses; seeconvert_positions_to_masses()position, a tuple of position segment labels; seeconvert_masses_to_positions()nltk, a string representation of segment positions; seeconvert_nltk_to_masses()
- An
Boundary Similarity (B)¶
This metric compares the correctness of boundary pairs between segmentations [Fournier2013].
Note
This is a recommended segmentation comparison metric for situations when there is no reference segmentation to compare against; see [Fournier2013].
-
segeval.boundary_similarity(segmentation_a, segmentation_b, **kwargs)¶ Parameters: segmentation_* (segmentation or Dataset) – Segmentation or dataset containing segmentations of a particular format; seeBoundaryFormat
-
segeval.boundary_similarity(dataset, **kwargs) Parameters: dataset ( Dataset) – Dataset of segmentations
-
segeval.boundary_similarity() Parameters: - boundary_format (
BoundaryFormatenum) – Segmentation format; defaultBoundaryFormat.mass - permuted (bool) – Use pairwise permutations v.s. combinations; default
False - one_minus (bool) – Return
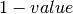 ; default
; default False - return_parts (bool) – Return tuples of numerators, demoninators, or other values comprising a metric; default
False - n_t (int) – See
boundary_edit_distance() - boundary_types (set) – Set of allowewable boundary types; default
set([1]) - weight (tuple) – Tuple of weighting functions, see Weighting Functions; default is scaling of substitution and transposition but not addition edits (
weight_a(),weight_s_scale(),weight_t_scale())
- boundary_format (
Segmentation Similarity (S)¶
Originally introduced in [FournierInkpen2012], this metric uses the revised boundary edit distance in [Fournier2013] and compares segmentations to provide the proportion of unedited potential boundary positions.
Warning
Prefer boundary_similarity() instead; see [Fournier2013].
-
segeval.segmentation_similarity(segmentation_a, segmentation_b, **kwargs)¶ For parameters see
boundary_similarity()
-
segeval.segmentation_similarity(dataset, **kwargs) For parameters see
boundary_similarity()
-
segeval.segmentation_similarity() For parameters see
boundary_similarity()
Boundary Edit Distance (BED)¶
An edit distance proposed in [Fournier2013] that operates upon boundaries to produce:
- Additions/deletion edits to model full misses,
- Transposition edits to model near misses, and
- Substitution edits to model boundary-type confusion.
For more details, see Section 3.1 of [Fournier2013b].
-
segeval.boundary_edit_distance(boundary_string_a, boundary_string_b, n_t=2)¶ Computes boundary edit distance between two boundary strings. Returns a list of Addition, Substitution, and Transposition edit sets.
Parameters: - boundary_string_a (tuple) – Boundary string to compare; produced by
boundary_string_from_masses() - boundary_string_b (tuple) – See boundary_string_a
- n_t (int) – Maximum distance (in potential boundary positions) that a transposition may span
- boundary_string_a (tuple) – Boundary string to compare; produced by
BED-based Confusion Matrix (BED-CM)¶
A confusion-matrix-formulation proposed in [Fournier2013] that uses BED to populate a matrix by using matches and scaled transpositions as correct classifications for boundary types, substitutions as confusion between boundary types, and additions/deletions as missing boundary types.
Note
This is a recommended segmentation comparison metric, when summarized by an information-retrieval metric such as precision(), recall(), fmeasure(), etc., for situations when there is a reference segmentation to compare against; see [Fournier2013].
-
segeval.boundary_confusion_matrix(hypothesis, reference, **kwargs)¶ Parameters: segmentation_* (segmentation) – Segmentation of a particular format; see BoundaryFormat
-
segeval.boundary_confusion_matrix(dataset, **kwargs) Parameters: dataset ( Dataset) – Dataset of segmentations
-
segeval.boundary_confusion_matrix(*args, **kwargs)
Weighting Functions¶
These functions are used by BED-based metrics to weight edit operations.
-
segeval.weight_a(additions)¶ Default unweighted weighting function for addition edit operations.
-
segeval.weight_s(substitutions, max_s, min_s=1)¶ Unweighted weighting function for substitution edit operations.
-
segeval.weight_s_scale(substitutions, max_s, min_s=1)¶ Default weighting function for substitution edit operations by the distance between ordinal boundary types.
-
segeval.weight_t(transpositions, max_n)¶ Unweighted weighting function for transposition edit operations.
-
segeval.weight_t_scale(transpositions, max_n)¶ Default weighting function for transposition edit operations by the distance that transpositions span.
Traditional Metrics¶
-
segeval.compute_window_size(reference, **kwargs)¶
Pk¶
Proposed in [BeefermanBerger1999], this segmentation comparison metric runs a window over a hypothesis and reference segmentation and counts those hypothesis windows whose ends are in differing segmentations that do not agree with the reference window as being in error. These errors are then summed over all windows.
Warning
Prefer boundary_similarity() instead; see [Fournier2013].
-
segeval.pk(hypothesis, reference, **kwargs)¶ Parameters: - hypothesis (segmentation or
Dataset) – Hypothetical, or automatically-generated, segmentation (or dataset of segmentations) of a particular format; seeBoundaryFormat - reference (segmentation or
Dataset) – Reference, or manually-created, segmentation (or dataset of segmentations) of a particular format; seeBoundaryFormat
- hypothesis (segmentation or
-
segeval.pk(dataset, **kwargs) Parameters: dataset ( Dataset) – Dataset of segmentations
-
segeval.pk() Parameters: - boundary_format (
BoundaryFormatenum) – Segmentation format; defaultBoundaryFormat.mass - permuted (bool) – Use pairwise permutations v.s. combinations; default
True - one_minus (bool) – Return
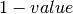 ; default
; default False - return_parts (bool) – Return tuples of numerators, demoninators, or other values comprising a metric; default
False - window_size (int) – Overriding window size – if not
None, this replaces the per-comparison window size computed usingcompute_window_size()as the window size used; defaultNone - fnc_round (function) – Rounding function used when computing window size, see
compute_window_size(); defaultround()
- boundary_format (
WindowDiff¶
Proposed in [PevznerHearst2002], this segmentation comparison metric is an adaptation of Pk which runs a window over a hypothesis and reference segmentation and counts those hypothesis windows with differing numbers of contained boundaries that do not agree with the reference window as being in error. These errors are then summed over all windows.
Warning
Prefer boundary_similarity() instead; see [Fournier2013].
-
segeval.window_diff(dataset, **kwargs) For parameters see
pk()
-
segeval.window_diff() For parameters see
pk()
Inter-coder Agreement Coefficients¶
Originally adapted in [FournierInkpen2012] from formulations provided by [ArtsteinPoesio2008], these have inter-coder agreement have been modified by [Fournier2013] to better suite the measurement of inter-coder agreement of segmentation boundaries using boundary_similarity() for actual agreement.
-
segeval.actual_agreement_linear()¶ Calculate actual (i.e., observed or
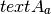 ), boundary agreement without accounting for chance, using [ArtsteinPoesio2008]‘s formulation as adapted by [Fournier2013].
), boundary agreement without accounting for chance, using [ArtsteinPoesio2008]‘s formulation as adapted by [Fournier2013].Parameters: - fnc_compare (function) – Segmentation comparison metric function to use; default
boundary_similarity() - boundary_format (
BoundaryFormatenum) – Segmentation format; defaultBoundaryFormat.mass - permuted (bool) – Use pairwise permutations v.s. combinations; default
False - one_minus (bool) – Return
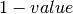 ; default
; default False - return_parts (bool) – Return tuples of numerators, demoninators, or other values comprising a metric; default
False - n_t (int) – See
boundary_edit_distance() - boundary_types (set) – Set of allowewable boundary types; default
set([1]) - weight (tuple) – Tuple of weighting functions, see Weighting Functions; default is scaling of substitution and transposition but not addition edits (
weight_a(),weight_s_scale(),weight_t_scale())
- fnc_compare (function) – Segmentation comparison metric function to use; default
-
segeval.fleiss_pi_linear(dataset, **kwargs)¶ Calculates Fleiss’
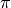 (or multi-
(or multi-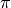 ), originally proposed in
[Fleiss1971], and is equivalent to Siegel and Castellan’s
), originally proposed in
[Fleiss1971], and is equivalent to Siegel and Castellan’s 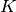 [SiegelCastellan1988]. For 2 coders, this is equivalent to Scott’s
[SiegelCastellan1988]. For 2 coders, this is equivalent to Scott’s 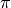 [Scott1955].
[Scott1955].For parameters see
actual_agreement_linear()
-
segeval.fleiss_kappa_linear(dataset, **kwargs)¶ Calculates Fleiss’
 (or multi-
(or multi- ), originally proposed in
[DaviesFleiss1982]. For 2 coders, this is equivalent to Cohen’s
), originally proposed in
[DaviesFleiss1982]. For 2 coders, this is equivalent to Cohen’s  [Cohen1960].
[Cohen1960].For parameters see
actual_agreement_linear()
-
segeval.artstein_poesio_bias_linear(dataset, **kwargs)¶ Artstein and Poesio’s annotator bias [ArtsteinPoesio2008].
For parameters see
actual_agreement_linear()
Format Conversion¶
These utility functions are used internally and provided to allow for the conversion between the supported segmentation formats (see BoundaryFormat).
-
segeval.boundary_string_from_masses(masses)¶ Creates a “boundary string”, or sequence of boundary type sets from a list of segment masses, e.g.,
[5,3,5]becomes[(),(),(),(),(1),(),(),(1),(),(),(),()].Parameters: masses (tuple) – Segmentation masses.
-
segeval.convert_positions_to_masses(positions)¶ Convert an ordered sequence of boundary position labels into a sequence of segment masses, e.g.,
[1,1,1,1,1,2,2,2,3,3,3,3,3]becomes[5,3,5].Parameters: segments (tuple) – Ordered sequence of which segments a unit belongs to. Deprecated since version 1.0.
-
segeval.convert_masses_to_positions(masses)¶ Converts a sequence of segment masses into an ordered sequence of section labels for each unit, e.g.,
[5,3,5]becomes[1,1,1,1,1,2,2,2,3,3,3,3,3].Parameters: masses (tuple) – Segment mass sequence.
-
segeval.convert_nltk_to_masses(string, boundary_symbol='1')¶ Convert an NLTK-formatted segmentation into masses, e.g.,
000001000100000becomes[5,3,5].For more information, see nltk.metrics.segmentation.
Parameters:
Data¶
These classes and functions deal with segmentation data representation and manipuation.
Model¶
These classes are used to model and store text (i.e., item) segmentations (i.e., codings).
-
class
segeval.Dataset(item_coder_data=None, properties=None, boundary_types=None, boundary_format='mass')¶ Represents a set of texts (i.e., items) that have been segmentations by coders.
-
copy()¶ Create a deep copy of the entire dataset object and properties.
-
-
class
segeval.Field¶ - An
enumwith options representing json fields when storing segmentations which include: segmentation_type, the type if segmentation; default isSegmentationType.linearitems, items with annotators and codings stored withincodings, annotators and codings stored within
- An
-
class
segeval.SegmentationType¶ - An
enumwith options representing segmentation structure types including: linear, linear segmentation
- An
Input/Output¶
These functions serialization and de-serialization segmentation datasets.
The recommended serialization format is JSON.
See also
-
segeval.input_linear_mass_tsv(filepath, delimiter='\t')¶ Takes a file path. Returns segmentation mass codings as a
Dataset.Parameters:
-
segeval.input_linear_mass_json(filepath)¶ Reads a file path. Returns segmentation mass codings as a
Dataset.Parameters: filepath ( str()) – Path to the mass file containing segment position codings.
-
segeval.output_linear_mass_json(filepath, dataset)¶ Takes a file path and
Datasetand serializes it as JSON.Parameters: filepath ( str()) – Path to the mass file containing segment position codings.
-
segeval.load_nested_folders_dict(containing_dir, filetype, dataset=None, prepend_item=[])¶ Loads TSV files from a file directory structure, which reflects the directory structure in nested
dict()with each directory name representing a key in thesedict().Parameters: